Genetic Methods of Species Identification
Reliability
Importance of Reliable Identification
Identifying an unknown specimen, putting a name to it, is often an important process. There may be economic, legal, or conservation imperatives for determining the identity. Often it is personal or scientific curiosity which drives us to ask "What is this?". Once we have an identification, a name, we are satisfied, but what if we are wrong? The consequences of error could be significant. Someone might be fined for having an illegally caught fish or a corporation might be sued for interfering with a rare species of bird. How can we be certain that the correct identification has been made?
Systematics and Classification
The science of organizing organisms into species, and then species into higher groups such as families and orders, is called systematics. This organised structure is called a classification or a taxonomy. Taxonomies have been constructed in the past on the basis of physical similarity, such as shape or colour, or even on the uses made of the organisms by people (e.g. herbs and medicines). Since Charles Darwin's "Origin of Species", there has been an ever increasing trend to build classifications which reflect the evolutionary relationships among organisms. That is, closely related species are placed together in the classification.
Identification Can be Difficult
An important part of systematics is determining how we can assign a specimen to one of a range of species. This is the process of identification. When physical characters are used, such as shape and colour, systematists try to find the combination of characteristics that allow you to identify the species reliably. These characteristics are not always obvious to the eye and may only be recognised by specialists who study a particular group of species. But, whether the characteristics be obvious, like an elephant's trunk, or very subtle, like the length of a fly's wing relative to its width, they provide important evidence of the specimen's species identity. This approach has been very successful and most identifications are made by such visual methods.
However, species are not always easily identified from distinctive external differences. Errors in identification can occur if these characters are difficult to interpret. There may be distinctive characters for only certain periods in the organism's life cycle. The characters may be fragile and easily damaged. Different species might not even have different, distinctive characters. When two populations of a species cease to interbreed, they do not necessarily develop different shape and colour, and such differences may not evolve for a very long time. Such pairs of species which are difficult or impossible to distinguish by external characters, are called cryptic (i.e., hidden) species. Until some way of recognizing these cryptic species is developed, members of them will be identified incorrectly.
Species can be quite variable in their characteristics, from one population to another. There have been many cases where systematists have mistakenly classified populations of a variable species as multiple species. Subsequently, when it has been recognised that these populations are actually interbreeding in nature, then the classification has been altered to combine them into one species.
So, any assessment of the reliability of molecular genetic methods of identification needs to be made in the context of less than perfect identification from external characteristics.
Reliable References
All of the molecular methods depend upon databases of reference sequences from reliably identified specimens. On a small scale, the Witness for the Whales subset of DNA Surveillance has been developed using sequences obtained from the specimens used to define the species, or from specimens identified by experts in whale biology. Many whales are very difficult to distinguish one from another, especially at sea, and many unreliable identifications have been made of a grey back slicing through the water. On a much larger scale, the Barcode of Life project aims to have every species represented by sequences derived from expertly identified specimens which have been saved and stored in archives. Some systems, such as GenBank which is searched using BLAST, are composed of publicly submitted sequences. Here there is no certainty that the specimens from which the sequences were derived have been identified correctly. Most specimens will be correctly identified, but rare or cryptic species might be mislabelled. The quality of the identification of the references has a direct impact on the reliability of the identification of an unknown specimen.
Testing Reliability
It is difficult to determine the reliability of a method of identification. You need the equivalent of the "blind taste test". That is, you give someone a specimen whose true identity is known, and the set of rules by which the identification should be made, and then determine how frequently they get the identity correct. If you are working with physical specimens and using characters associated with shape and colour, then you could ask how often do two experts disagree. That, however, is as much a test of their expertise as it is of the accuracy of the rules they must use to make the identification. We have no independent knowledge of the truth, and only know how others have approached the problem.
The genetic methods of identification are somewhat different, in that they provide explicit and relatively unambiguous methods for making an identification. Once the sequence is determined then computer programmes can apply a set of decision rules to make an identification. Again it is difficult to do the tests using real specimens because you are always comparing a human judgement against one done with the computer. When they disagree, how can we be certain which is correct?
One approach which removes the confounding effect of human judgement is to use computer simulation. With this approach you first build an artificial phylogenetic tree containing several imaginary species. Then you use software to simulate the evolution of DNA along this tree. It does this by randomly adding changes at a rate in proportion to the length of each branch. This will produce a set of realistic DNA sequences with several from specimens from each species.
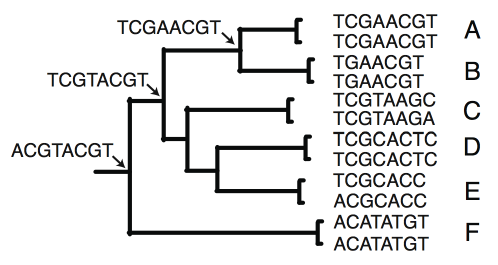
In the tree above we see the simulated sequence at the base of the tree and at some of the internal nodes. At the tips, there are two sequences per species. This shows just a short example section of DNA. Normally you would use longer sequences many hundreds of bases long.
Then you take some of these sequences away, to be used as your test sequences, and leave the rest to be the reference sequences. Now you can try to identify the species to which each test sequence belongs, and then compare the result with the known answer.
The simulation approach allows you to control for factors like human skill and judgement. However, if the approach that you used to simulate the evolution of the sequences was not very realistic then the results of your test will have little meaning.
Results of Simulation Tests
Simulations have been done using a lot of different combinations of variables. In general these experiments show:
- The three methods, BLAST, DNA Barcoding and phylogenetic methods work equally well.
- A major cause of mistaken identification is if the true species is not in the reference dataset. This could happen if there are cryptic species or if a species, perhaps a rare one, has not been sampled and included among the references.
- Another major cause of misidentification is if the genetic differences between species is not significantly greater than the genetic variation within species. Shortly after speciation occurs, the new species will be very similar genetically. There may be strong selection for them to become different at the characters that really matter, traits associated with reproduction such as those involved in courtship, mate attraction or pollinator attraction. However, the genetic regions that are commonly used in species identification tend not to be associated with any biologically important traits. Consequently young species may not be distinguishable.
Conclusions
Most of the time, with most species, these methods will give reliable identifications. This is a young science, and we are still in a discovery phase. There remain many species for which we lack genetic sequences. As time passes more reference sequences will be available, making these methods more reliable. However, our ability to distinguish young species will always limit the reliability of these techniques.
